Authors:
(1) Jinge Wang, Department of Microbiology, Immunology & Cell Biology, West Virginia University, Morgantown, WV 26506, USA;
(2) Zien Cheng, Department of Microbiology, Immunology & Cell Biology, West Virginia University, Morgantown, WV 26506, USA;
(3) Qiuming Yao, School of Computing, University of Nebraska-Lincoln, Lincoln, NE 68588, USA;
(4) Li Liu, College of Health Solutions, Arizona State University, Phoenix, AZ 85004, USA and Biodesign Institute, Arizona State University, Tempe, AZ 85281, USA;
(5) Dong Xu, Department of Electrical Engineer and Computer Science, Christopher S. Bond Life Sciences Center, University of Missouri, Columbia, MO 65211, USA;
(6) Gangqing Hu, Department of Microbiology, Immunology & Cell Biology, West Virginia University, Morgantown, WV 26506, USA ([email protected]).
Table of Links
4. Biomedical Text Mining and 4.1. Performance Assessments across typical tasks
4.2. Biological pathway mining
5.1. Human-in-the-Loop and 5.2. In-context Learning
6. Biomedical Image Understanding
7.1 Application in Applied Bioinformatics
7.2. Biomedical Database Access
7.2. Online tools for Coding with ChatGPT
7.4 Benchmarks for Bioinformatics Coding
8. Chatbots in Bioinformatics Education
9. Discussion and Future Perspectives
4. BIOMEDICAL TEXT MINING
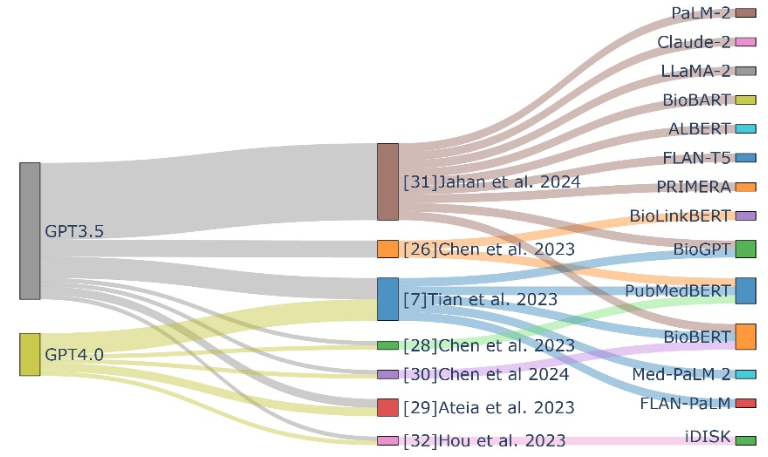
For biomedical text mining with ChatGPT, we first summarized works that evaluate the performance of ChatGPT in various biomedical text mining tasks and compared it to stateof-the-art (SOTA) models (Figure 2). Then, we explored how ChatGPT has been used to reconstruct biological pathways and prompting strategies used to improve performance.
4.1. PERFORMANCE ASSESSMENTS ACROSS TYPICAL TASKS
Biomedical text mining tasks typically include name entity recognition, relation extraction, sentence similarity, document classification, and question answering. Chen, Sun [26] assessed ChatGPT-3.5 across 13 publicly available benchmarks. While its performance in question answering closely matched SOTA models like PubmedBERT[27], ChatGPT-3.5 showed limitations in other tasks, with similar observations made for ChatGPT-4[7, 28, 29]. Extensions to sentence classification and reasoning revealed that ChatGPT was inferior to SOTA pretrained models like BioBERT[30]. These studies highlight the limitations of ChatGPT in some specific domains of biomedical text mining where domain-optimized language models excel. Nevertheless, when the training sets with task-specific annotations are not sufficient, zero-shot LLMs, including ChatGPT-3.5 outperform SOTA finetuned biomedical models[31].
Biomedical Knowledge Graphs (BKGs) have emerged as a novel paradigm for managing large-scale, heterogeneous biomedical knowledge from expert-curated sources. Hou, Yeung [32] evaluated ChatGPT’s capability on question and answering tasks using topics collected from the “Alternative Medicine” subcategory on “Yahoo! Answers” and compare to the Integrated Dietary Supplements Knowledge Base (iDISK)[33]. While ChatGPT-3.5 showed comparable performance to iDISK, ChatGPT-4 was superior to both. However, when tasked to predict drug or dietary supplement repositioned for Alzheimer's Disease, ChatGPT primarily responded with candidates already in clinical trials or existing literature. Moreover, ChatGPT’s efforts to establish associations between Alzheimer's Disease and hypothetical substances were less than impressive. This highlights ChatGPT's limitations in performing novel discoveries or establishing new entity relationships within BKGs.
ChatGPT's underperformance in some specific text mining tasks against SOTA models or BKGs identifies areas for enhancement; On the other hand, finetuning LLMs, although beneficial, remains out of reach for most users due to the high computational demand. Therefore, techniques like prompt engineering, including one/few-shot in-context learning and CoT, can be more practical to improve LLM efficiency in text mining tasks[26, 28, 30, 34]. For instance, incorporating examples with CoT reasoning enhances the performances over both zero-shot (no example) and plain examples in sentence classification and reasoning tasks[30] as well as knowledge graph reconstruction from literature titles[35]. However, simply increasing the number of examples does not always correlate with better performance[28, 30]. This underscores another challenge in optimizing LLMs for specialized text mining tasks, necessitating more efficient prompting strategies to ensure consistent reliability and stability.
4.2. BIOLOGICAL PATHWAY MINING
Another emerging application of biomedical text mining from LLMs is to build biological pathways. The ChatPathway project[36] assessed ChatGPT-3.5's intrinsic capability to predict biochemical reactions and map out metabolic or regulatory pathways based on given reactants and enzymes. Using KEGG annotations as benchmarks, it reported modest prediction accuracies: 24.1% for biochemical reactions, 24.6% to 42.3% for metabolic pathways, and 20.1% to 39.9% for regulatory pathways. Azam, Chen [37] conducted a broader assessment of mining gene interactions and biological pathways across 21 LLMs, including seven Application Programming Interface (API)-based and 14 open-source models. ChatGPT-4 and Claude-Pro emerged as leaders, though they only achieved F1 scores less than 0.5 for gene relation predictions and a Jaccard index less than 0.3 for pathway predictions. Another evaluation work on retrieving protein-protein interaction (PPI) from sentences reported a modest F1 score for both GPT-3.5 and GPT-4 for base prompts[38]. All the studies underscore the inherent challenges generic LLMs face in delineating gene relationships and constructing complex biological pathways from biomedical text without prior knowledge or specific training.
Nevertheless, the capabilities of ChatGPT in knowledge extraction and summarization present promising avenues for database curation support. Tiwari, Matthews [39] explored its utility in the Reactome curation process, notably in identifying potential proteins for established pathways and generating comprehensive summaries. For the case study on the circadian clock pathway, ChatGPT proposed 13 new proteins, five of which were supported by the literature but overlooked in traditional manual curation. When summarizing pathway from multiple literature extracts, ChatGPT struggled to resolve contradictions, but gained improved performance when inputs contained in-text citations. Similarly, the use of ChatGPT for annotating long non-coding RNAs in the EVLncRNAs 3.0 database[40] faces issues with inaccurate citations. Both works demonstrate the challenge of direct use of ChatGPT in assisting in database curation.
Supplementing ChatGPT with domain knowledge or literature has been shown to mitigate some of its intrinsic limitations. The inclusion of a protein dictionary in prompts improves performance for GPT-3.5 and GPT-4 in PPI task[38]. Chen, Li [41] augmented ChatGPT with literature abstracts to identify genes involved in arthrofibrosis pathogenesis. Similarly, Fo, Chuah [42] supplied GPT-3.5 with plant biology abstracts to uncover over 400,000 functional relationships among genes and metabolites. This domain knowledge/literature-backed approach enhances the reliability of chatbots in text generation by reducing AI hallucination[43, 44].
Addressing LLMs' intrinsic limitations can also involve sophisticated prompt engineering. Chen, Gao [45] introduced an iterative prompt optimization procedure to boost ChatGPT's accuracy in predicting genegene interactions, utilizing KEGG pathway database as a benchmark. Initial tests without prompt enhancements showed a performance decline along with ChatGPT’s upgrades from March to July in 2023, but the strategic role and few-shot prompts significantly countered this trend. The iterative optimization process, which employed the tree-of-thought methodology[46], achieved notable improvements in precision and F1 scores. These experiments demonstrate the value of strategic prompt engineering in aligning LLM outputs with complex biological knowledge for better performance.
This paper is available on arxiv under CC BY 4.0 DEED license.